Submitted by ve10 on
What little time I had last week, not addressing various end-of-semester issues, I spent writing the folding code. I’m happy to report that not only am I just a few functions away from a fully functional solver, but I very much like the form it has taken. Once again I have to thank Alex Alemi, this time for pushing me to go as far as defining structs of structs and other C arcana that, remarkably, add a degree of transparency.
Because I have no results to report — I had to cut short my coding to write the final exam for Analytical Mechanics — I will instead describe the odd way the solver represents the structure of a protein.
Previously I explained that with divide and concur one works with multiple copies of the variables, such that constraints may easily be applied in each copy. Sudoku uses four copies, where the first three are exactly the ones anyone thinks about, because leaving a space blank is not an option you normally consider. This only comes up when the 9x9 grid of digits is replaced by a 9x9x9 grid of 0’s and 1’s. Now all the constraints are of the form “exactly one number in this set is a 1, the others are 0”, including the constraint that each column “above” the 9x9 grid is of this form.
But how does divide and concur work with continuous variables, such as we have with proteins? The animation below shows the algorithm solving a disk packing problem:
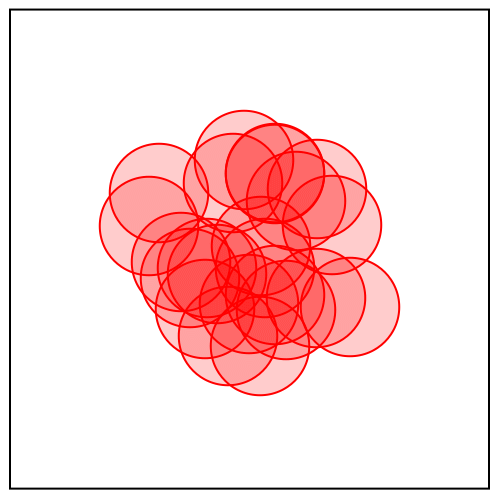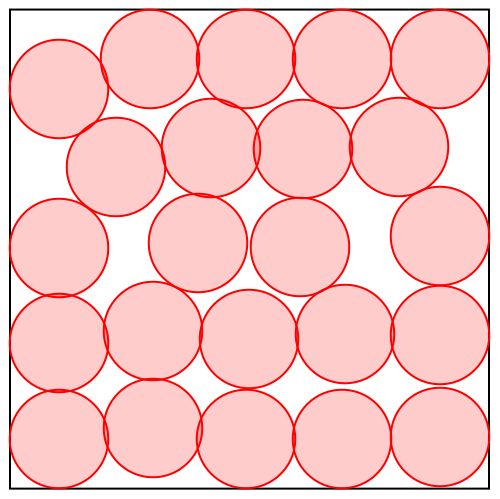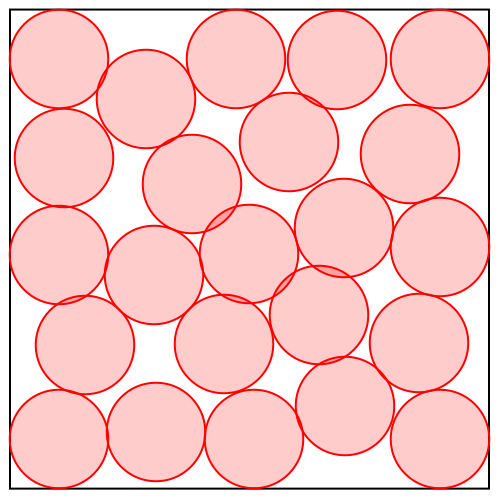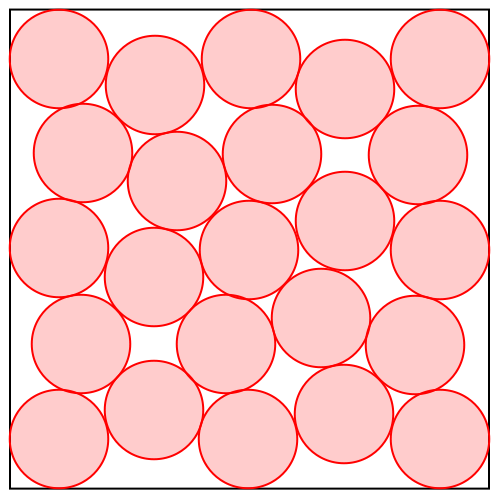
It looks like there are 23 disks, always with some pairs overlapping until the last frame, when the solution is found. The algorithm works with many more disks than this: 23x22 or 506 in all. Each of the 23 disks you see in the animation actually has 22 copies that happen to be at the same position. These are outputs of the concur projection.
The divide projection does different things to the different copies. Consider say “disk 1” in the animation and its 22 copies. One of these copies you can think of as “the copy of disk 1 that avoids overlap with disk 2”. Another copy is “the copy of disk 1 that avoids overlap with disk 3”, etc. The no-overlap constraint between disks 1 and 2 applies only to “the copy of disk 1 that avoids overlap with disk 2” and “the copy of disk 2 that avoids overlap with disk 1”, and that is also the only constraint that applies to these copies.
Projecting to the no-overlap constraint is easy because it is applied to only pairs of disk-copies that are oblivious to all others. If they are already not overlapping there is no change; otherwise they are moved the minimum distance so this is true.
How many copies do we need of a protein on which we want to impose many litemotif constraints? Recall the protein is represented by just the α-carbon atoms in its backbone. If the primary sequence has R residues, then we need R backbone atoms. There will also be a certain number of solvent oxygen atoms that participate in the litemotif constraints. Say there are S of these.
Ideally we would like to be able to impose a litemotif constraint on each of the R-3 (overlapping) sets of four consecutive residues along the backbone. The actual number will be lower, owing to a combination of two things. First, by the simple fact of bad luck our primary sequence my contain 4-residue subsequences that happen to be underrepresented in the PDB, and we should not take the few available samples as an exhaustive survey of structures. Second, our multiple side-chain contact criterion for litemotifhood may not apply to certain parts of the backbone, in particular the “loops” between better packed alpha-helix and beta-sheet domains. In any case, the actual number of litemotif constraints C is likely to be somewhat smaller than R-3.
We have R+S atom positions and C constraints, each of which is allocated its own copy of all the positions. The solver therefore acts on (R+S)xC vector variables. Here’s how to think about these copies. Copy #1 is associated with residues 1234, copy #2 with residues 2345, and so on, skipping those residue subsequences for which we don’t have sufficient samples in the PDB.
The constraint that applies to copy #1 is mostly about backbone atoms 1234. Projecting to the constraint goes as follows: replace atoms 1234, and one other atom among the remaining backbone or solvent atoms, by a 5-atom litemotif structure with matching residues (including ‘O’ for solvent contact). Here “replace” means “replace by a RMSD minimizing rotated-translated litemotif”. There will be dozens of litemotifs with matching residues, and a prospective replacement is computed for each one. The replacement having the smallest RMSD is the one that will be used. When the projection to constraint #1 is completed, only backbone atoms 1234 and one other atom (backbone or solvent) will have a new position in copy #1. The rule for determining the “other atom” is not ad hoc in the least, but a true distance minimizing projection to a PDB-certified configuration. The other constraints (on 2345, 3456, etc.) are computed likewise, and just on their copies.
Concur does two things. First and foremost it gives all copies of an atom the same position (the mean of the copies). However, without straying from the projection idea, it can do more. As an example, I can have a constraint that all backbone atoms must lie within some compact region and all except a minority of solvent atoms must lie outside that region. Now, after I compute the mean position of the copies, I check whether this position is in the allowed region; if not, I move it to the nearest point on the boundary of the allowed region.
Once again I have run out of steam without having had a chance to discuss the potential pitfalls in this scheme. The plan was to foreshadow various mechanisms of disaster and thereby lessen the sting of embarrassment, should that be the eventual outcome. But on second thought, my time is better spent writing code.
